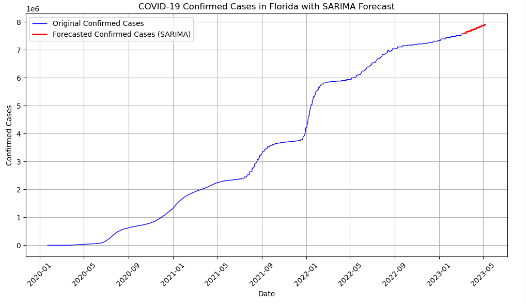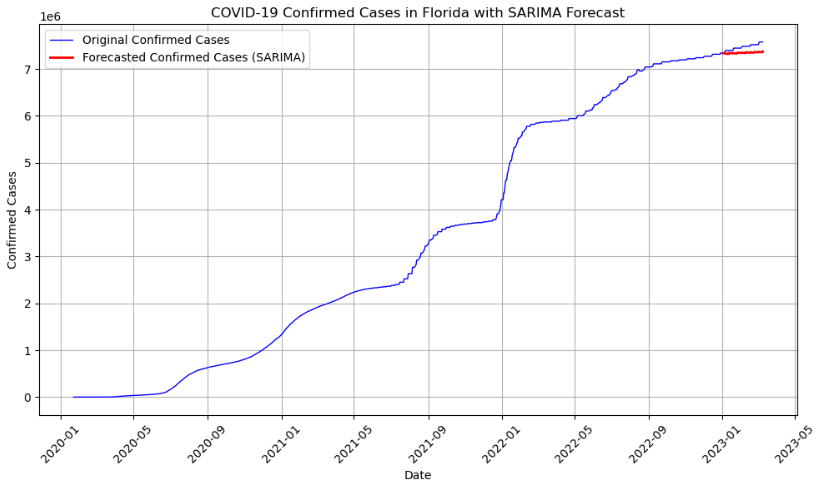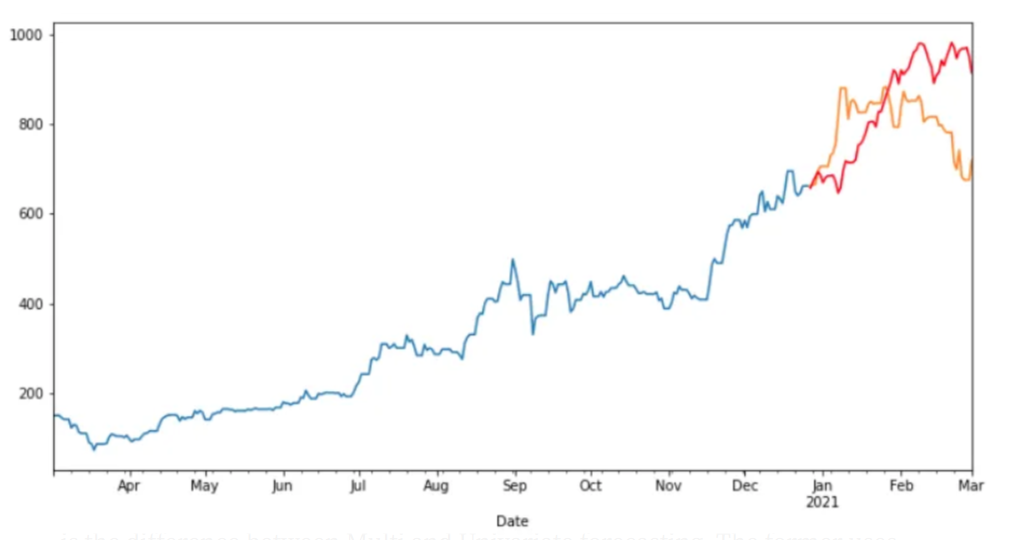
Source: An Introduction to Time Series Analysis with ARIMA, Towards Data Science
Abstract
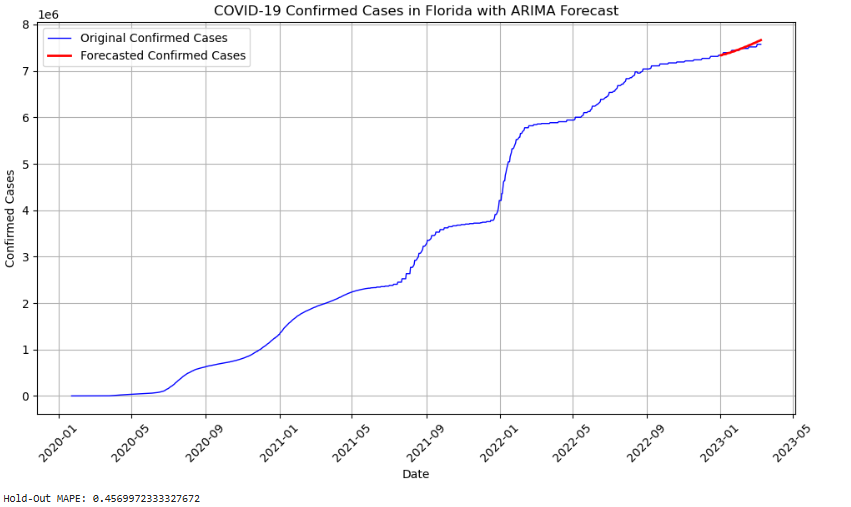
This study explores time series forecasting techniques using ARIMA, SARIMA, and Bayesian Structural Time Series (BSTS) models on two distinct datasets: the COVID-19 dataset from the Johns Hopkins CSSEGISandData repository and the CO₂ emissions data from Mauna Loa, Hawaii, provided by NOAA. The primary objective is to apply machine learning techniques to predict future data patterns and to evaluate the efficiency of each model, comparing their performance against each other For large datasets, such as COVID-19, both ARIMA and SARIMA models demonstrated rapid processing capabilities; however, SARIMA introduced additional noise due to explicit seasonality, impacting accuracy. Both models produced forecasts with higher variability and were outperformed by the BSTS model, which demonstrated lower Root Mean Squared Error (RMSE) and Hold-Out Mean Absolute Percentage Error (MAPE) but with greater computational complexity causing slower processing time.
In contrast, the smaller Mauna Loa CO₂ dataset, characterized by smoother seasonal patterns, allowed for more precise forecasting. Among the tested models, the BSTS model achieved the highest accuracy with an RMSE of 0.765 and a MAPE of 0.161%, outperforming ARIMA and SARIMA, which had RMSE values of 12.55 and 13.38, respectively. This analysis highlights the effectiveness of the BSTS model for stable, seasonally predictable datasets, while ARIMA-based models may struggle with complex, volatile data like the COVID-19 dataset. These findings underscore the importance of selecting models that align with the dataset’s characteristics to optimize forecasting accuracy.
Introduction
Anything that is observed sequentially over time is a time series. In the data, I will look at regular intervals of time. Time series forecasting is a statistical method used to predict future values based on past results. The simplest models focus solely on the variable being forecast, ignoring external factors like marketing efforts or economic shifts, and instead extrapolating existing trends and seasonality. Specifically in this capstone, I will be exploring univariate models ARIMA/SARIMA and the Bayesian STS.
The use of time series analysis gained recognition since 1970 when the Box-Jenkins method popularized ARIMA models to find the best fit of different time series models based on past data. However, the concept of analyzing sequential data over time is not new. As the world increasingly embraces data-driven solutions to forecast future industry outcomes, time series analysis has become vital in numerous fields.
Currently, efforts to perfect the science of forecasting is important as the higher the accuracy of a time series based on past data and events, the more essential the tool becomes for modern organizations as accurate forecasts support decision-making across various timeframes—whether short-term, medium-term, or long-term, depending on the specific application. Key considerations in forecasting include seasonality, trends, and external fluctuations, which play a significant role in shaping predictions. These specifically will be explored more as I deal with the data sets of this project. These forecasts are integral for planning, resource allocation, and adjusting strategies in response to upcoming movements.
The main goal of my paper is to explore and discuss the concept and use real life datasets in order to see how time series analysis can use its historical data and leverage practical and meaningful conclusions. In this paper, we will also be discussing any possible limitations that the different time series models have and what some suggestions are to improve our predictions and possibly overcome these limitations.
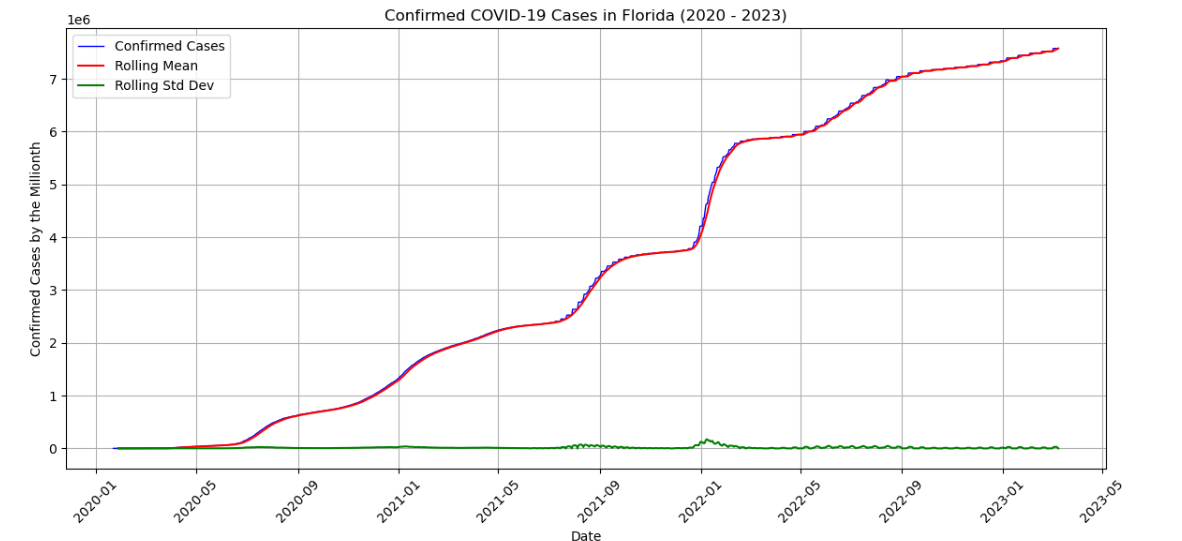
In this capstone, I will explore the Johns Hopkins CSSEGISandData COVID-19 Repository, which tracks global COVID-19 cases and Our World in Data (OWID), which provides comprehensive COVID-19 datasets across various indicators datasets to test the univariate models ARIMA/SARIMA and Bayesian STS. Using the same model structures, I will also be exploring the Forecasting CO₂ Emissions in Mauna Loa, Hawaii. This dataset is provided by the Global Monitoring Laboratory within NOAA. The goal of this exploration is to apply time series analysis to forecast the future spread of COVID-19, seeing which models are the strongest to do such a task, and to use those same models to appropriately forecast the increasing trend of the CO₂ emission rates. These two different datasets and applications are to show the strength in forecasting in multiple real-life situations. This is important in data science and statistics, particularly in public health and global policy as creating accurate forecasts can allow policymakers a better direction on what should be applied and when.
To achieve the goals of my capstone, ARIMA/SARIMA will be used for capturing linear trends and seasonal effects, and Bayesian STS for incorporating uncertainty and providing probabilistic forecasts. The project will begin with an exploratory data analysis (EDA) to visualize the time series data, identify patterns, and check for seasonality or trends. After cleaning and preprocessing the data, I will test both models using Python on the COVID-19 and CO₂ datasets. The results will then be compared to assess which approach yields the most accurate and insightful forecasts.
Time Series Forecasting Models
In different papers and studies, it is easy to see that there are a multitude of models to explore and chose from, depending on what is being questioned or asked from by the researcher and data. Most of the models can be seen following into different categories. Traditional statistical modeling, which includes a moving average, exponential smoothing, and autoregressive (ARIMA). These are seen as linear functions taking past observations to make sound predictions. Second category of model type is nonlinear models. This will not be covered in my project, but the main difference lies in the complexity and non-proportional relationships in the data. An example of this can be LSTM, ANN, NAR models. The second main model type that I will be covering is the Bayesian Structural Time Series (BSTS). This model type is considered linear, since it relies on the state-space models, but can handle much more complexity such as trend, seasonality, and regression components. So, although it is linear, the approach is more flexible by adding prior distributions allowing for uncertainty estimations. In this capstone, these two ARIMA/SARIMA and Bayesian STS will be explore in depth.
ARIMA Model
To begin, throughout this project I will be utilizing the ARIMA model as a traditional method of time series forecasting. ARIMA stands for Autoregressive integrated moving average and it predicts future values with past values. It utilizes lagged moving averages to smooth the time series data. The ARIMA model is a univariate model that can be broken down into two different models, AR and MA, which will be define below.
The ARIMA model is defined by the parameters \( (p, d, q) \), where:
- \( p\) is the number of lag observations (autoregressive terms).
- \( d\) is the number of times the data must be differenced to make it stationary.
- \( q\) is the number of lagged forecast errors (moving average terms) [1].
The ARIMA model is composed of two submodels, the AR (Autoregressive) and the MA (Moving Average) model, described mathematically as follows:
\[Y(AR)_t = \alpha + \beta_1 Y(AR)_{t-1} + \beta_2 Y(AR)_{t-2} + \cdots + \beta_p Y(AR)_{t-p}\] (1)
where \( Y(AR)_t \) are the lags of the series, \( \beta \) are the lag coefficients estimated by the model, and \( \alpha \) is the model’s intercept term.
\[Y(MA)_t = \epsilon_t + \theta_1 \epsilon_{t-1} + \theta_2 \epsilon_{t-2} + \cdots + \theta_q \epsilon_{t-q}\] (2)
where \( Y(MA)_t \) depends only on the lagged forecast errors, \( \theta \) are the parameters of the model, and \( \epsilon \) are unrelated error terms.
The mathematical expression of the ARIMA model, as a combination of AR and MA models, is:
\[Y_t = \alpha + \sum_{i=1}^{p} \beta_i Y_{t-i} + \epsilon_t - \sum_{j=1}^{q} \theta_j \epsilon_{t-j}\] (3)
Where \( Y_t \) and \( \epsilon_t \) represent the time series and the random error at time \( t \), \( \beta \) and \( \theta \) are the coefficients of the model.
When estimating the \( p\) and \( q\) parameters of the AR and the MA models, plots from the ACF function and the PACF functions are used. Just to summarize, the autocorrelation function, ACF, is a function that shows the correlation between the observed of the \(t \) time and the observation at the previous times. This function indicates the autocorrelation coefficient, which is the measurement of the correlation between observations at different times. PACF, or partial autocorrelation function differs because it is the correlation between two random variables after the influence of confounding variable is removed [7].
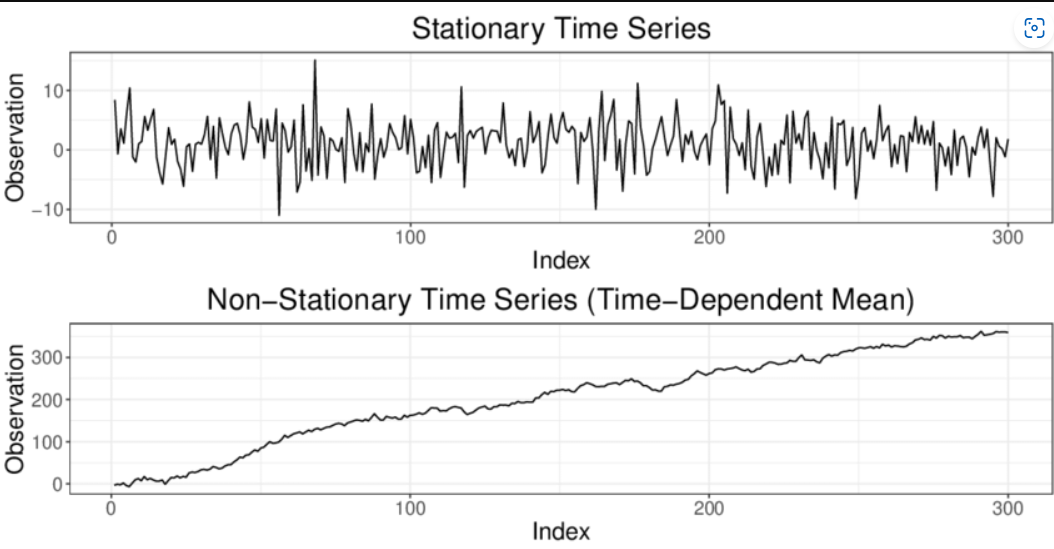
Source: Classification of Time Series
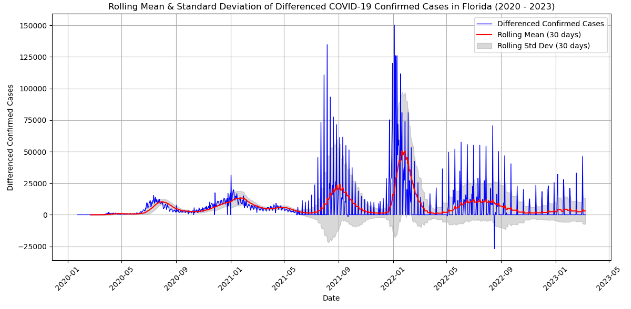
Prior to finding the ACF/PACF, it is obligated for us to know if the data that is obtained is stationary or nonstationary. This is important because if the data is not stationary, the data cannot perform the ARIMA model accurately. Therefore, in order to determine if the model is stationary, it can be visualized or the Dickey-Fuller Test can be performed to determine the classification of the time series data.
When differencing is applied and observed, the \(p\) order deals with the AR model and it is based on the significant spikes from the PACF plot. The MA model, \(q\) order is determined if the ACF plots have sharp cut-off after lags. If there has been any differencing applied to the model, \(d\) is determined by the order of the differencing (i.e. how many times the differencing has been applied).
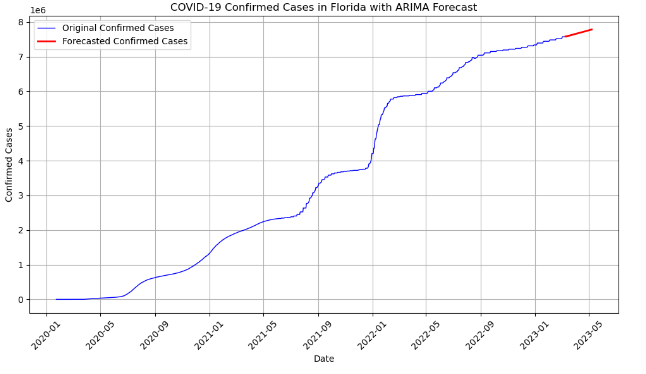
After the strongest combination of the \( (p, d, q) \) have been selected, it is possible to then run the ARIMA model and see the results. At times, it can be difficult to physically pick out the correct combination. If this becomes the case, there are machine learning approaches to choose the appropriate orders.
Bayesian Structural Time Series Model
Fundamentally, this theorem is unique from ARIMA due to differences in the interpretation of what “probability” means. We will explore criticisms later, but it is important to remember this distinction when understanding the model differences. Bayesian inference is a method of statistical inference where available knowledge about parameters in a statistical model is updated with information from observed data. A Guide to Bayesian Inference.
Bayesian inference provides a powerful framework to update beliefs as new data becomes available. It allows us to calculate the probability of an event occurring, given prior knowledge and new evidence. A simplified version of Bayes’ theorem looks like this:
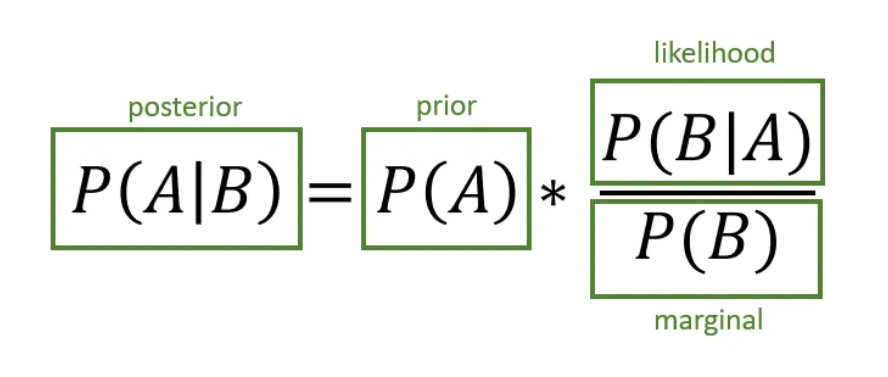
Bayesian Time Series Forecasting
Referring to the image explaining the visualization, it breaks down the components as follows:
- Prior (P(A)): The initial belief about the probability of event A, before new evidence is considered.
- Likelihood (P(B | A)): The probability of observing evidence B given that event A has occurred.
- Marginal (P(B)): The total probability of observing the evidence across all possible events.
- Posterior (P(A | B)): The updated belief about event A given the new evidence B.
This framework is fundamental in Bayesian time series analysis, allowing models to update predictions as new data arrives. The posterior distribution represents the estimation of a parameter, ensuring the model reflects both the past trends and the latest observations.
Source: Created based on Bayes' theorem principles, adapted for better interpretability.
Now, for Bayesian Structural Time-Series Models, the time series can be broken down into four basic models depending on the following: a level, a local trend, seasonal impacts, and an error term. Structural Time Series models can be defined by the following:
Equation (4) and Equation (5) represent the structure of the model. Equation (4) is trying to understand the distribution of the \( Y_t \)’s of the data, while Equation (5) is closely related to understanding the distribution of the parameters. It provides the evolution model for the parameters and how they change over time. Because the vector evolves through randomness, it describes the distribution of the hidden parameters at the next step. This is crucial because for Bayesian inference to hold, updating the beliefs of the parameters helps incorporate both the priors and the observed data.
Criticism and Limitations
The key distinction between the ARIMA and Bayesian methods lies in their underlying theoretical perspectives: frequentist vs. Bayesian. In the frequentist approach (as used in ARIMA models), parameters are treated as fixed values that do not change once estimated. The data is analyzed under the assumption that these parameters have unique, fixed points.
In contrast, the Bayesian approach views parameters as random variables that can evolve over time, with uncertainty captured through prior distributions. This flexibility allows the Bayesian method to incorporate prior knowledge about the parameter’s movement and update these beliefs as new data becomes available, resulting in what is known as posterior distributions.
This difference introduces a potential limitation: if the prior distribution is inaccurate or misinformed, it can negatively affect the model’s output. While prior information can enhance predictions, it can also become a burden if the assumptions are flawed or if prior knowledge conflicts with new observations. Therefore, Bayesian models offer both advantages and risks: they excel when reliable prior knowledge is available but can be inaccurate if the priors introduce bias into the model.
Next, time series analysis inherently faces criticism because it attempts to predict the future—data that has not yet materialized—with as much certainty as possible. When practicing time series forecasting, it is crucial to not only critique the model’s inference but also incorporate evaluation methods into the decision-making process, given the inherent uncertainty of projections.
While cross-validation techniques are still applicable and recommended for time series models, specific approaches such as leave-one-out (LOO) cross-validation can pose challenges. For time series, LOO can be problematic when the goal is to estimate predictive performance for future time points, as removing an observation disrupts the temporal dependencies in the data. This is particularly problematic from a Bayesian perspective, where each time point informs the next. Ignoring these dependencies undermines the theoretical foundation of time series models, which rely on the structure of sequential observations.